
[ Home | Writing | Research | Music | Miscellaneous ]
During my time in the Boxer Lab in 2018, I embarked on a crystallographic journey to uncover some mysteries about the photophysics of fluorescent proteins.

Many years ago, scientists discovered the first fluorescent proteins in glowing jellyfish. Since then, people have created all sorts of variations of fluorescent proteins for cellular imaging. The particular type of fluorescent protein that I studied is called a photoswitchable fluorescent protein. It behaves like a togglable light-bulb. When you shine a blue 488nm laser on the protein, it stops fluorescing and becomes dark; when you shine a purple 405nm laser on it, it starts fluorescing again. By exploiting this photoswitching property in clever ways, you can take higher-resolution images of cells (known as super-resolution microscopy). But to design better photoswitching proteins, it's helpful to have some sense of the physical principles behind their function.
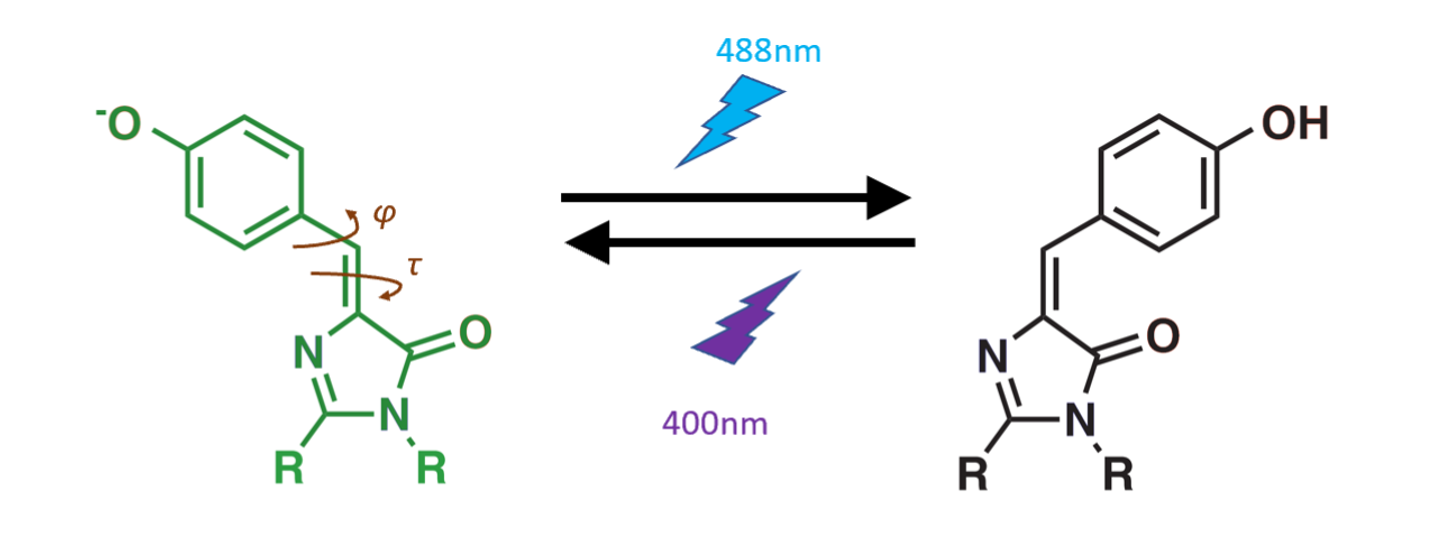
In my project, I wanted to understand the mechanism of the photoswitching process. We know that the on-to-off behavior occurs because the light-emitting part of the protein (known as the chromophore) changes shape from cis to trans when it absorbs a photon of light. But we didn't know the exact trajectory of the atoms as they rearrange from a cis conformation to a trans conformation. So we wanted to elucidate (pun intended) the photo-isomerization mechanism of these photoswitching proteins.

As part of my research, I crystallized these fluorescent proteins to determine their structure via X-ray diffraction. On the left, I've shown the mesmerizing nanoscale structure of crystalline protein. You can see the protein units closely packed against each other in a regular, repeating lattice. As you'd imagine, such a closely packed environment is quite different than the typical environment inside of a cell.
While solving crystal structures, we made the surprising discovery that the geometry of the protein's internal chromophore pocket could be perturbed by the packing interactions between the protein units of the crystal. This sort of finding makes you really question whether crystal structures of proteins reflect their structures in vivo!
Keep an eye out for a publication later this year, where this fascinating finding will be reported in more detail.
Update Sep 2019: The paper is now published in JACS, hooray!
Stay tuned for more...
[ Home | Writing | Research | Music | Miscellaneous ]